REGISTRATION – Current version: 2.4, released December 2024.
Note: Atomic Interaction analysis is no longer maintained in DL_ANALYSER. A dedicated software called D_ATA (Atom Typer and Analyser) has been developed to carry out interaction analysis.
DL_ANALYSER is a utility software tool for DL_POLY molecular dynamics simulation package.
It can carry out a variety of post analysis work on the DL_POLY’s HISTORY trajectory files. The program contains a collection of unified analysis tools that can produce a variety of results concurrently in a single-read through a collection of trajectory files. The nature and type of results to produce can be controlled in one single DL_ANALYSER’s control (dl_analyser.control) file. The analysis options can also be applied on a selection of atoms specified by the users.
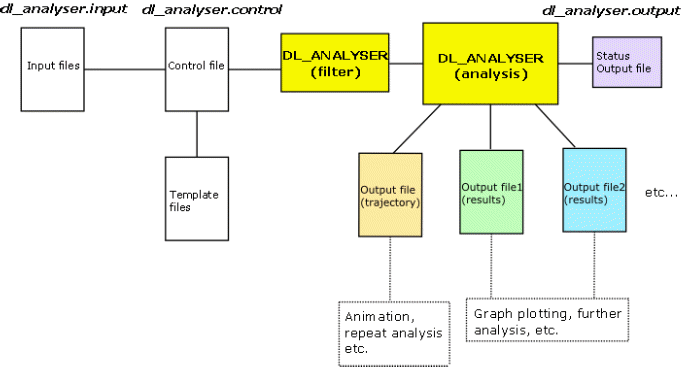
Features
- Input structure format (ether in uncompress or gzip): HISTORY trajectory file, CONFIG, STATIS, PDB and xyz. Also reads DL_MESO binary HISTORY file.
- Write out trajectories in the following format: PDB, xyz.
- Flexible, user-selectable analysis options: atom-based, molecule-based, and index-based.
- Analysis Sections: Structural, Dynamical, Defect, Surface Sputtering, Biological and Atomic Interactions.
- Unique feature (DANAI): Capable to identify, quantify and annotate interactions at atomic scales.
DANAI – DL_ANALYSER Notation for Atomic Interaction, a standard, searchable notation to describe atomic interactions. The feature is only partially implemented in DL_ANALYSER.
Relevant links
DL_FIELD – utility software to setup force field models that uses DL_F Notation to describe atoms for DANAI analysis
DL_ANALYSER tutorial – available at DL_SDG site.
References:
If you use DL_ANALYSER in your work, please use the following references:
- C.W. Yong, I.T. Todorov ‘DL_ANALYSER Notation for Atomic Interactions (DANAI): A natural annotation system for molecular interactions, using ethanoic acid liquid as a test case’ Molecules 23, 36 (2018)
- I. Rosbottom, C.W. Yong, D.L. Geatches, R.B. Hammond, I.T. Todorov and K.J. Roberts ‘The integrated DL_POLY/DL_FIELD/DL_ANALYSER software platform for molecular dynamics simulations for exploration of the synthonic interactions in saturated benzoic acid/hexane solutions’ Mole. Simul. 47, 257-272 (2021)
